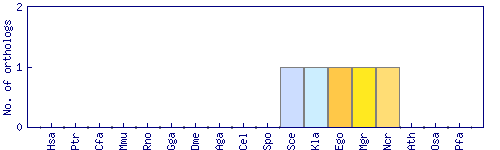
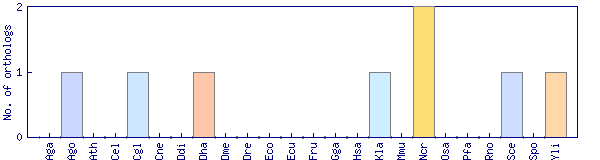
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | FLC2 | ||||||||||
| SGD link | S000000049 | ||||||||||
| Alternative ID | YAL053W | ||||||||||
| Description | Putative FAD transporter; required for uptake of FAD into endoplasmic reticulum; involved in cell wall maintenance | ||||||||||
| Synonyms | YAL053W | ||||||||||
| Datasets |
|