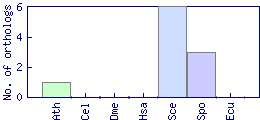
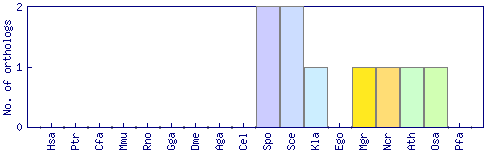
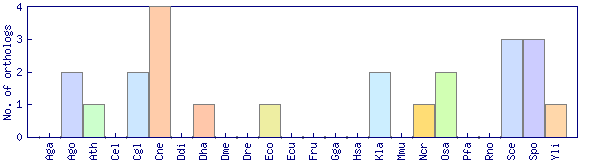
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | FUR4 | ||||||||||
| SGD link | S000000225 | ||||||||||
| Alternative ID | YBR021W | ||||||||||
| Description | Uracil permease, localized to the plasma membrane; expression is tightly regulated by uracil levels and environmental cues | ||||||||||
| Synonyms | YBR021W | ||||||||||
| Datasets |
|