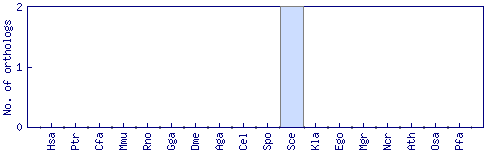
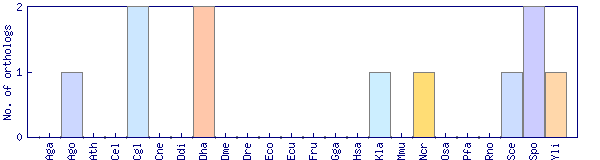
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | ECM33 | ||||||||||
| SGD link | S000000282 | ||||||||||
| Alternative ID | YBR078W | ||||||||||
| Description | GPI-anchored protein of unknown function, has a possible role in apical bud growth; GPI-anchoring on the plasma membrane crucial to function; phosphorylated in mitochondria; similar to Sps2p and Pst1p | ||||||||||
| Synonyms | YBR078W | ||||||||||
| Datasets |
|