| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | RXT2 | ||||||||||
| SGD link | S000000299 | ||||||||||
| Alternative ID | YBR095C | ||||||||||
| Description | Subunit of the histone deacetylase Rpd3L complex; possibly involved in cell fusion and invasive growth | ||||||||||
| Synonyms | YBR095C | ||||||||||
| Datasets |
|
| YOGY Home | YOGY Help |
|
| HomoloGene cluster | Phylogenetic pattern |
| 38410 | 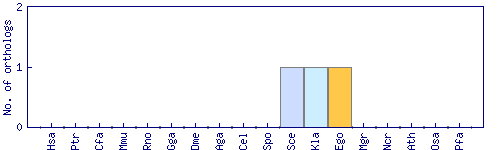 |
| Species | Gene ID | NCBI link | UniProt link |
| Saccharomyces cerevisiae (Sce) | RXT2 (S000000299) | GI:37362619 | |
| Kluyveromyces lactis (Kla) | KLLA0B11990g | GI:50304227 | |
| Eremothecium gossypii (Ego) | AGOS_ADL313W | GI:45187560 |
| GO ID | GO term | GO aspect | Evidence codes and associations | ||||
| GO:0005634 | nucleus | C |
| ||||
| GO:0033698 | Rpd3L complex | C |
| ||||
| GO:0070210 | Rpd3L-Expanded complex | C |
| ||||
| GO:0004407 | histone deacetylase activity | F |
| ||||
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | P |
| ||||
| GO:0000747 | conjugation with cellular fusion | P |
| ||||
| GO:0001403 | invasive growth in response to glucose limitation | P |
| ||||
| GO:0016575 | histone deacetylation | P |
| ||||
| GO:0031939 | negative regulation of chromatin silencing at telomere | P |
|
| OrthoMCL cluster | Phylogenetic pattern |
| OG1_15295 | 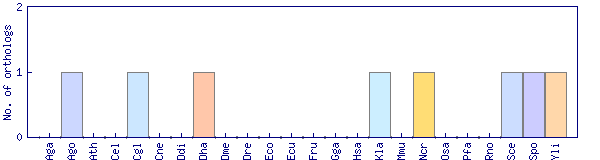 |
| Species | OrthoMCL sequence | Model organism protein page | UniProt link |
| Ashbya gossypii (Ago) | ago1428 | ADL313W | |
| Candida glabrata (Cgl) | cgl2435 | CAGL0H08481g | |
| Debaryomyces hansenii (Dha) | dha3166 | DEHA0D19393g | |
| Kluyveromyces lactis (Kla) | kla1061 | KLLA0B11990g | |
| Neurospora crassa (Ncr) | ncr5811 | NCU05979.2 | |
| Saccharomyces cerevisiae (Sce) | sce354 | YBR095C (S000000299) | |
| Schizosaccharomyces pombe (Spo) | spo452 | SPBC428.06c | O94355 |
| Yarrowia lipolytica (Yli) | yli2196 | YALI0C11517g |
| GO ID | GO term | GO aspect | Evidence codes and associations | ||||||||
| GO:0005634 | nucleus | C |
| ||||||||
| GO:0032221 | Rpd3S complex | C |
| ||||||||
| GO:0033698 | Rpd3L complex | C |
| ||||||||
| GO:0070210 | Rpd3L-Expanded complex | C |
| ||||||||
| GO:0003674 | molecular_function | F |
| ||||||||
| GO:0004407 | histone deacetylase activity | F |
| ||||||||
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | P |
| ||||||||
| GO:0000747 | conjugation with cellular fusion | P |
| ||||||||
| GO:0001403 | invasive growth in response to glucose limitation | P |
| ||||||||
| GO:0006338 | chromatin remodeling | P |
| ||||||||
| GO:0016575 | histone deacetylation | P |
| ||||||||
| GO:0031939 | negative regulation of chromatin silencing at telomere | P |
|
Curated yeast ortholog results:
| S. pombe systematic ID | S. cerevisiae systematic ID |
| SPBC428.06c (O94355) | YBR095C (S000000299) |
| GO ID | GO term | GO aspect | Evidence codes and associations | ||||||||
| GO:0005634 | nucleus | C |
| ||||||||
| GO:0032221 | Rpd3S complex | C |
| ||||||||
| GO:0033698 | Rpd3L complex | C |
| ||||||||
| GO:0070210 | Rpd3L-Expanded complex | C |
| ||||||||
| GO:0003674 | molecular_function | F |
| ||||||||
| GO:0004407 | histone deacetylase activity | F |
| ||||||||
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | P |
| ||||||||
| GO:0000747 | conjugation with cellular fusion | P |
| ||||||||
| GO:0001403 | invasive growth in response to glucose limitation | P |
| ||||||||
| GO:0006338 | chromatin remodeling | P |
| ||||||||
| GO:0016575 | histone deacetylation | P |
| ||||||||
| GO:0031939 | negative regulation of chromatin silencing at telomere | P |
|
| YOGY Home | YOGY Help |
| s.khadayate@ucl.ac.uk |