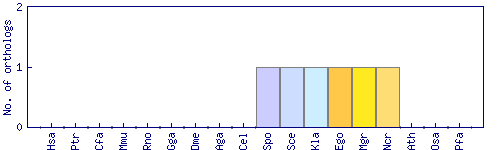
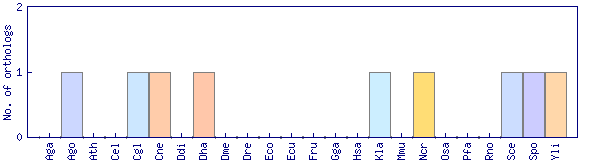
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | MET8 | ||||||||||
| SGD link | S000000417 | ||||||||||
| Alternative ID | YBR213W | ||||||||||
| Description | Bifunctional dehydrogenase and ferrochelatase, involved in the biosynthesis of siroheme, a prosthetic group used by sulfite reductase; required for sulfate assimilation and methionine biosynthesis | ||||||||||
| Synonyms | YBR213W | ||||||||||
| Datasets |
|