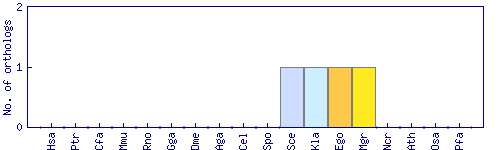
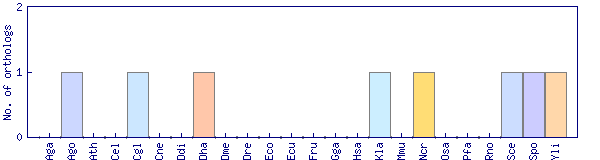
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | HPC2 | ||||||||||
| SGD link | S000000419 | ||||||||||
| Alternative ID | YBR215W | ||||||||||
| Description | Subunit of the HIR complex, a nucleosome assembly complex involved in regulation of histone gene transcription; mutants display synthetic defects with subunits of FACT, a complex that allows passage of RNA Pol II through nucleosomes | ||||||||||
| Synonyms | YBR215W | ||||||||||
| Datasets |
|