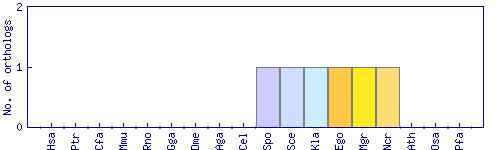
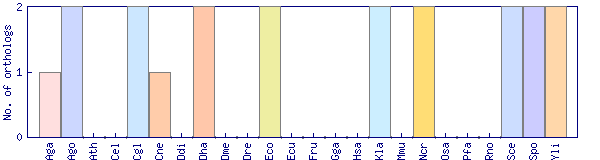
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | ARO4 | ||||||||||
| SGD link | S000000453 | ||||||||||
| Alternative ID | YBR249C | ||||||||||
| Description | 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase, catalyzes the first step in aromatic amino acid biosynthesis and is feedback-inhibited by tyrosine or high concentrations of phenylalanine or tryptophan | ||||||||||
| Synonyms | YBR249C | ||||||||||
| Datasets |
|