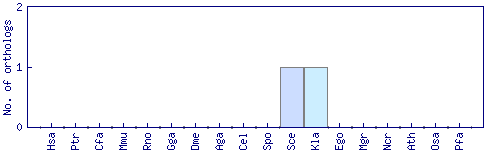
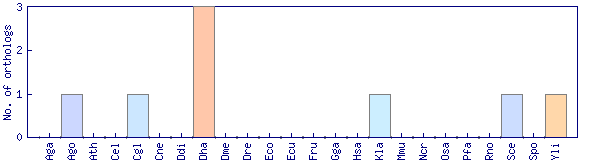
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | MTC4 | ||||||||||
| SGD link | S000000459 | ||||||||||
| Alternative ID | YBR255W | ||||||||||
| Description | Protein of unknown function, required for normal growth rate at 15 degrees C; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm in a punctate pattern; mtc4 is synthetically sick with cdc13-1 | ||||||||||
| Synonyms | YBR255W | ||||||||||
| Datasets |
|