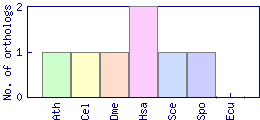
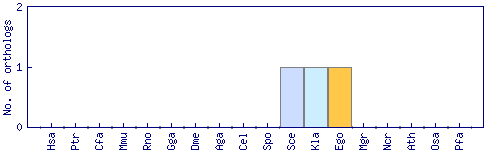
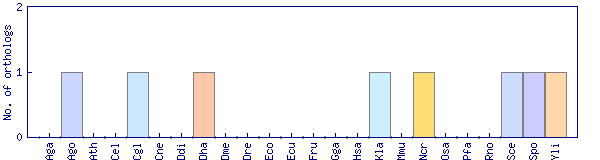
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | APM3 | ||||||||||
| SGD link | S000000492 | ||||||||||
| Alternative ID | YBR288C | ||||||||||
| Description | Mu3-like subunit of the clathrin associated protein complex (AP-3); functions in transport of alkaline phosphatase to the vacuole via the alternate pathway | ||||||||||
| Synonyms | YBR288C | ||||||||||
| Datasets |
|