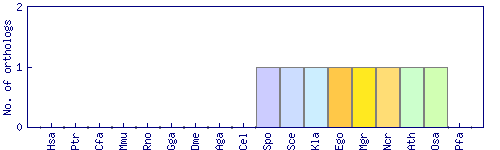
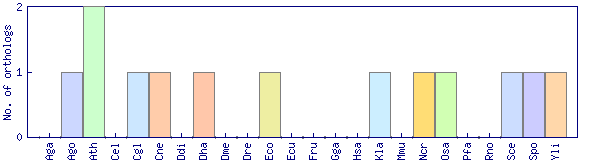
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | HIS4 | ||||||||||
| SGD link | S000000535 | ||||||||||
| Alternative ID | YCL030C | ||||||||||
| Description | Multifunctional enzyme containing phosphoribosyl-ATP pyrophosphatase, phosphoribosyl-AMP cyclohydrolase, and histidinol dehydrogenase activities; catalyzes the second, third, ninth and tenth steps in histidine biosynthesis | ||||||||||
| Synonyms | YCL030C | ||||||||||
| Datasets |
|