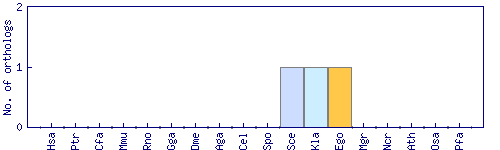
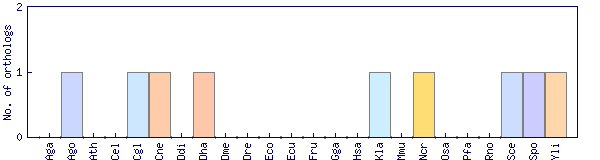
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | MRC1 | ||||||||||
| SGD link | S000000566 | ||||||||||
| Alternative ID | YCL061C | ||||||||||
| Description | S-phase checkpoint protein required for DNA replication; interacts with and stabilizes Pol2p at stalled replication forks during stress, where it forms a pausing complex with Tof1p and is phosphorylated by Mec1p; protects uncapped telomeres | ||||||||||
| Synonyms | YCL061C | ||||||||||
| Datasets |
|