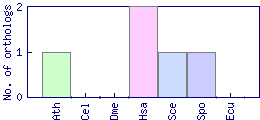
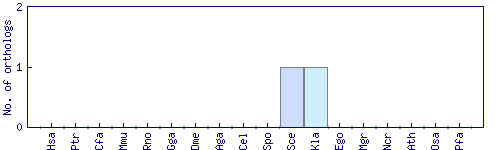
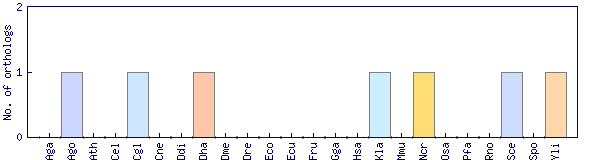
| GO ID | GO term | GO aspect | Evidence codes and associations |
| GO:0000176 | nuclear exosome (RNase complex) | C | |
| GO:0000177 | cytoplasmic exosome (RNase complex) | C | |
| GO:0005654 | nucleoplasm | C | |
| GO:0005730 | nucleolus | C | |
| GO:0005737 | cytoplasm | C | |
| GO:0003674 | molecular_function | F | |
| GO:0000467 | exonucleolytic trimming to generate mature 3'-end
of 5.8S rRNA from tricistronic rRNA transcript
(SSU-rRNA, 5.8S rRNA, LSU-rRNA) | P | |
| GO:0034427 | nuclear-transcribed mRNA catabolic process,
exonucleolytic, 3'-5' | P | |
| GO:0043628 | ncRNA 3'-end processing | P | |
| GO:0070478 | nuclear-transcribed mRNA catabolic process, 3'-5'
exonucleolytic nonsense-mediated
decay | P | |
| GO:0070481 | nuclear-transcribed mRNA catabolic process,
non-stop decay | P | |
| GO:0070651 | nonfunctional rRNA decay | P | |
| GO:0071035 | nuclear polyadenylation-dependent rRNA catabolic
process | P | |
| GO:0071038 | nuclear polyadenylation-dependent tRNA catabolic
process | P | |
| GO:0071042 | nuclear polyadenylation-dependent mRNA catabolic
process | P | |
| GO:0071051 | polyadenylation-dependent snoRNA 3'-end
processing | P | |
| GO ID | GO term | GO aspect | Evidence codes and associations |
| GO:0000176 | nuclear exosome (RNase complex) | C | |
| GO:0000177 | cytoplasmic exosome (RNase complex) | C | |
| GO:0005654 | nucleoplasm | C | |
| GO:0005730 | nucleolus | C | |
| GO:0005737 | cytoplasm | C | |
| GO:0003674 | molecular_function | F | |
| GO:0000467 | exonucleolytic trimming to generate mature 3'-end
of 5.8S rRNA from tricistronic rRNA transcript
(SSU-rRNA, 5.8S rRNA, LSU-rRNA) | P | |
| GO:0034427 | nuclear-transcribed mRNA catabolic process,
exonucleolytic, 3'-5' | P | |
| GO:0043628 | ncRNA 3'-end processing | P | |
| GO:0070478 | nuclear-transcribed mRNA catabolic process, 3'-5'
exonucleolytic nonsense-mediated
decay | P | |
| GO:0070481 | nuclear-transcribed mRNA catabolic process,
non-stop decay | P | |
| GO:0070651 | nonfunctional rRNA decay | P | |
| GO:0071035 | nuclear polyadenylation-dependent rRNA catabolic
process | P | |
| GO:0071038 | nuclear polyadenylation-dependent tRNA catabolic
process | P | |
| GO:0071042 | nuclear polyadenylation-dependent mRNA catabolic
process | P | |
| GO:0071051 | polyadenylation-dependent snoRNA 3'-end
processing | P | |
| GO ID | GO term | GO aspect | Evidence codes and associations |
| GO:0000176 | nuclear exosome (RNase complex) | C | |
| GO:0000177 | cytoplasmic exosome (RNase complex) | C | |
| GO:0005634 | nucleus | C | |
| GO:0005654 | nucleoplasm | C | |
| GO:0005730 | nucleolus | C | |
| GO:0005737 | cytoplasm | C | |
| GO:0005829 | cytosol | C | |
| GO:0000175 | 3'-5'-exoribonuclease activity | F | |
| GO:0003674 | molecular_function | F | |
| GO:0000467 | exonucleolytic trimming to generate mature 3'-end
of 5.8S rRNA from tricistronic rRNA transcript
(SSU-rRNA, 5.8S rRNA, LSU-rRNA) | P | |
| GO:0006364 | rRNA processing | P | |
| GO:0034427 | nuclear-transcribed mRNA catabolic process,
exonucleolytic, 3'-5' | P | |
| GO:0043628 | ncRNA 3'-end processing | P | |
| GO:0070478 | nuclear-transcribed mRNA catabolic process, 3'-5'
exonucleolytic nonsense-mediated
decay | P | |
| GO:0070481 | nuclear-transcribed mRNA catabolic process,
non-stop decay | P | |
| GO:0070651 | nonfunctional rRNA decay | P | |
| GO:0071035 | nuclear polyadenylation-dependent rRNA catabolic
process | P | |
| GO:0071038 | nuclear polyadenylation-dependent tRNA catabolic
process | P | |
| GO:0071042 | nuclear polyadenylation-dependent mRNA catabolic
process | P | |
| GO:0071051 | polyadenylation-dependent snoRNA 3'-end
processing | P | |