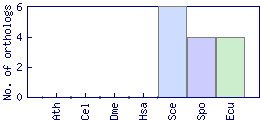
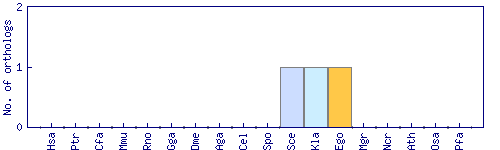
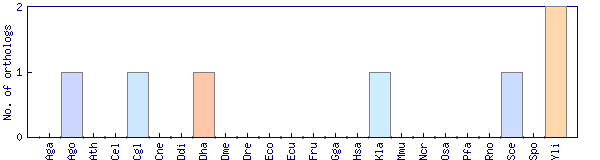
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | RRT12 | ||||||||||
| SGD link | S000000641 | ||||||||||
| Alternative ID | YCR045C | ||||||||||
| Description | Probable subtilisin-family protease with a role in formation of the dityrosine layer of spore walls; localizes to the spore wall and also the nuclear envelope and ER region in mature spores | ||||||||||
| Synonyms | YCR045C | ||||||||||
| Datasets |
|