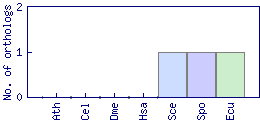
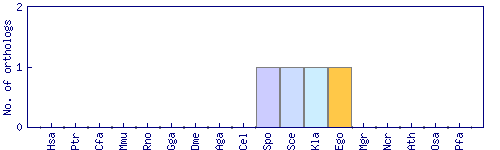
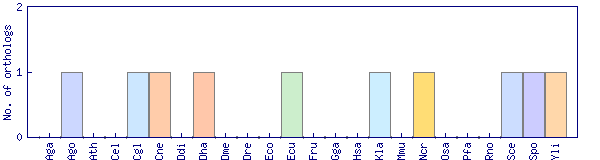
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | ATG15 | ||||||||||
| SGD link | S000000664 | ||||||||||
| Alternative ID | YCR068W | ||||||||||
| Description | Lipase required for intravacuolar lysis of autophagic bodies and Cvt bodies; targeted to intravacuolar vesicles during autophagy via the multivesicular body (MVB) pathway | ||||||||||
| Synonyms | YCR068W | ||||||||||
| Datasets |
|