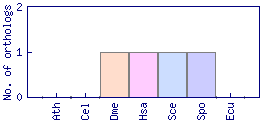
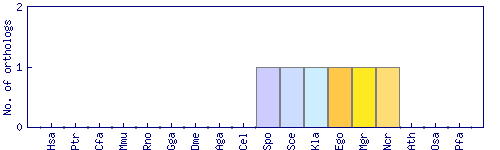
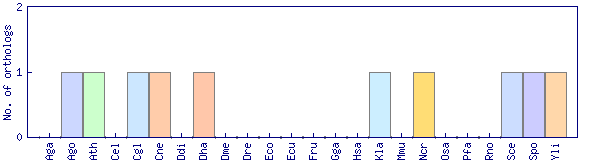
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | PAT1 | ||||||||||
| SGD link | S000000673 | ||||||||||
| Alternative ID | YCR077C | ||||||||||
| Description | Topoisomerase II-associated deadenylation-dependent mRNA-decapping factor; also required for faithful chromosome transmission, maintenance of rDNA locus stability, and protection of mRNA 3'-UTRs from trimming; functionally linked to Pab1p | ||||||||||
| Synonyms | YCR077C | ||||||||||
| Datasets |
|