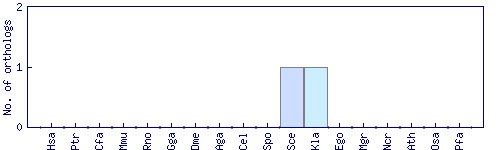
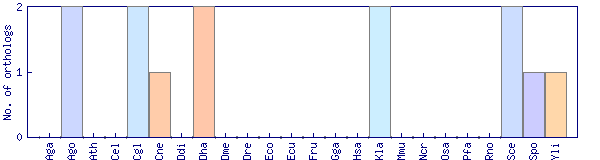
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | KNH1 | ||||||||||
| SGD link | S000002207 | ||||||||||
| Alternative ID | YDL049C | ||||||||||
| Description | Protein with similarity to Kre9p, which is involved in cell wall beta 1,6-glucan synthesis; overproduction suppresses growth defects of a kre9 null mutant; required for propionic acid resistance | ||||||||||
| Synonyms | YDL049C | ||||||||||
| Datasets |
|