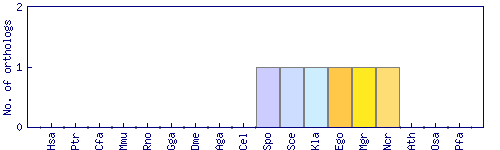
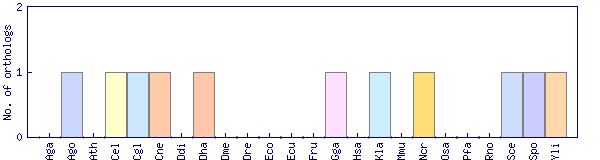
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | LRG1 | ||||||||||
| SGD link | S000002399 | ||||||||||
| Alternative ID | YDL240W | ||||||||||
| Description | Putative GTPase-activating protein (GAP) involved in the Pkc1p-mediated signaling pathway that controls cell wall integrity; appears to specifically regulate 1,3-beta-glucan synthesis | ||||||||||
| Synonyms | YDL240W | ||||||||||
| Datasets |
|