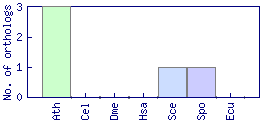
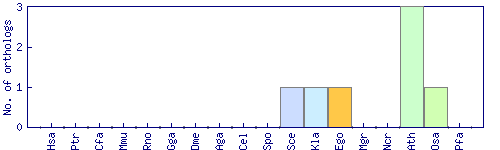
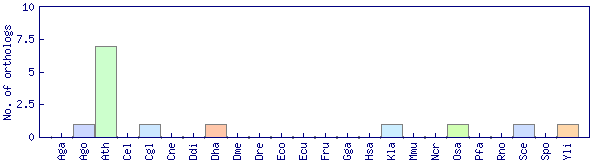
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | TRP1 | ||||||||||
| SGD link | S000002414 | ||||||||||
| Alternative ID | YDR007W | ||||||||||
| Description | Phosphoribosylanthranilate isomerase that catalyzes the third step in tryptophan biosynthesis; in 2004, the sequence of TRP1 from strain S228C was updated by changing the previously annotated internal STOP (TAA) to serine (TCA) | ||||||||||
| Synonyms | YDR007W | ||||||||||
| Datasets |
|