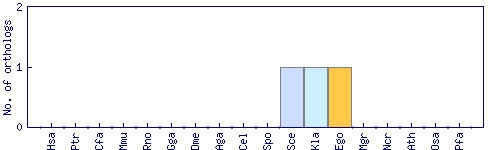
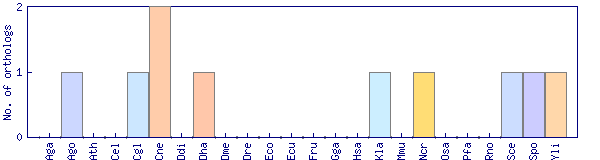
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | REG1 | ||||||||||
| SGD link | S000002435 | ||||||||||
| Alternative ID | YDR028C | ||||||||||
| Description | Regulatory subunit of type 1 protein phosphatase Glc7p, involved in negative regulation of glucose-repressible genes | ||||||||||
| Synonyms | YDR028C | ||||||||||
| Datasets |
|