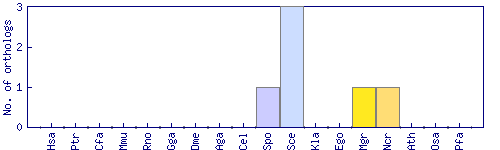
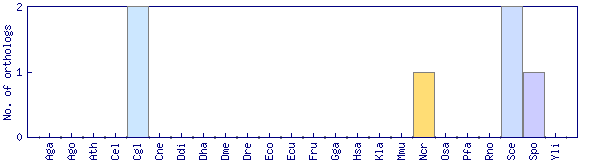
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | MRH1 | ||||||||||
| SGD link | S000002440 | ||||||||||
| Alternative ID | YDR033W | ||||||||||
| Description | Protein that localizes primarily to the plasma membrane, also found at the nuclear envelope; the authentic, non-tagged protein is detected in mitochondria in a phosphorylated state; has similarity to Hsp30p and Yro2p | ||||||||||
| Synonyms | YDR033W | ||||||||||
| Datasets |
|