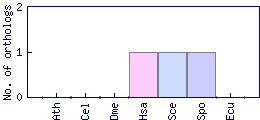
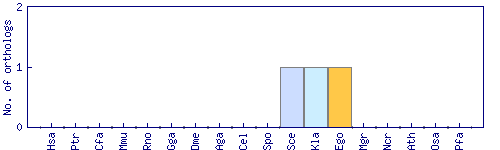
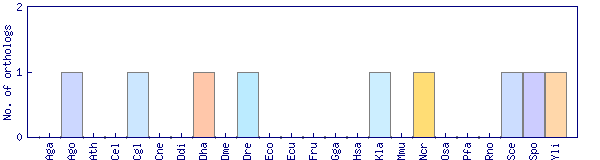
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | PAA1 | ||||||||||
| SGD link | S000002478 | ||||||||||
| Alternative ID | YDR071C | ||||||||||
| Description | Polyamine acetyltransferase; acetylates polyamines (e.g. putrescine, spermidine, spermine) and also aralkylamines (e.g. tryptamine, phenylethylamine); may be involved in transcription and/or DNA replication | ||||||||||
| Synonyms | YDR071C | ||||||||||
| Datasets |
|