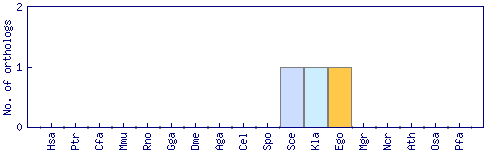
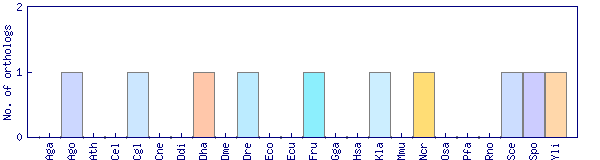
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | NBP2 | ||||||||||
| SGD link | S000002569 | ||||||||||
| Alternative ID | YDR162C | ||||||||||
| Description | Protein involved in the HOG (high osmolarity glycerol) pathway, negatively regulates Hog1p by recruitment of phosphatase Ptc1p the Pbs2p-Hog1p complex, found in the nucleus and cytoplasm, contains an SH3 domain that binds Pbs2p | ||||||||||
| Synonyms | YDR162C | ||||||||||
| Datasets |
|