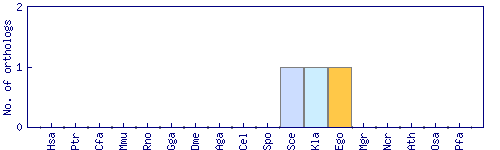
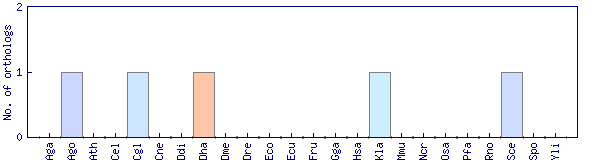
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | RAV2 | ||||||||||
| SGD link | S000002610 | ||||||||||
| Alternative ID | YDR202C | ||||||||||
| Description | Subunit of RAVE (Rav1p, Rav2p, Skp1p), a complex that associates with the V1 domain of the vacuolar membrane (H+)-ATPase (V-ATPase) and promotes assembly and reassembly of the holoenzyme | ||||||||||
| Synonyms | YDR202C | ||||||||||
| Datasets |
|