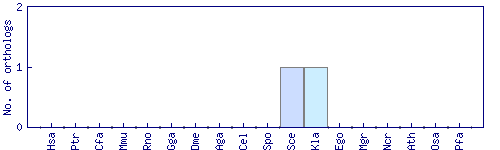
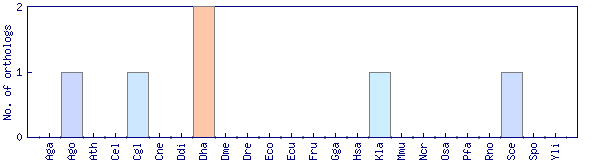
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | UME6 | ||||||||||
| SGD link | S000002615 | ||||||||||
| Alternative ID | YDR207C | ||||||||||
| Description | Key transcriptional regulator of early meiotic genes, binds URS1 upstream regulatory sequence, couples metabolic responses to nutritional cues with initiation and progression of meiosis, forms complex with Ime1p, and also with Sin3p-Rpd3p | ||||||||||
| Synonyms | YDR207C | ||||||||||
| Datasets |
|