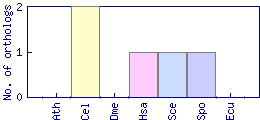
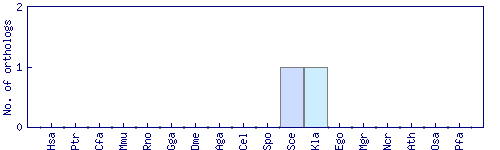
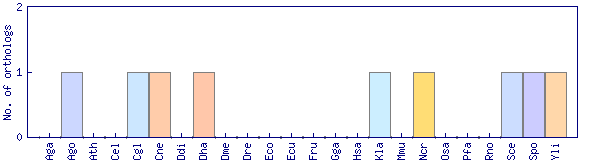
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | RAD9 | ||||||||||
| SGD link | S000002625 | ||||||||||
| Alternative ID | YDR217C | ||||||||||
| Description | DNA damage-dependent checkpoint protein, required for cell-cycle arrest in G1/S, intra-S, and G2/M; transmits checkpoint signal by activating Rad53p and Chk1p; hyperphosphorylated by Mec1p and Tel1p; potential Cdc28p substrate | ||||||||||
| Synonyms | YDR217C | ||||||||||
| Datasets |
|