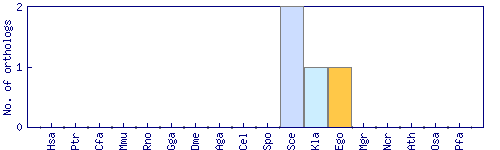
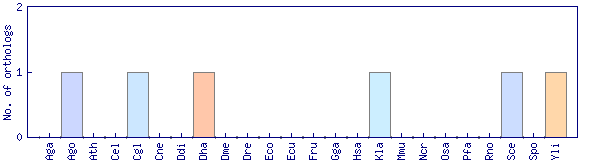
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | MTH1 | ||||||||||
| SGD link | S000002685 | ||||||||||
| Alternative ID | YDR277C | ||||||||||
| Description | Negative regulator of the glucose-sensing signal transduction pathway, required for repression of transcription by Rgt1p; interacts with Rgt1p and the Snf3p and Rgt2p glucose sensors; phosphorylated by Yck1p, triggering Mth1p degradation | ||||||||||
| Synonyms | YDR277C | ||||||||||
| Datasets |
|