| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | PRO1 | ||||||||||
| SGD link | S000002708 | ||||||||||
| Alternative ID | YDR300C | ||||||||||
| Description | Gamma-glutamyl kinase, catalyzes the first step in proline biosynthesis | ||||||||||
| Synonyms | YDR300C | ||||||||||
| Datasets |
|
| YOGY Home | YOGY Help |
|
| KOG name | Phylogenetic pattern (and link to KOGs with this pattern) |
| KOG1154 | ACDHYP- |
| KOGs classification | KOGs description |
| METABOLISM | Gamma-glutamyl kinase |
| GO ID | GO term | GO aspect | Evidence codes and associations | ||||||||||||||||||
| GO:0005737 | cytoplasm | C |
| ||||||||||||||||||
| GO:0009507 | chloroplast | C |
| ||||||||||||||||||
| GO:0016020 | membrane | C |
| ||||||||||||||||||
| GO:0003674 | molecular_function | F |
| ||||||||||||||||||
| GO:0004349 | glutamate 5-kinase activity | F |
| ||||||||||||||||||
| GO:0016301 | kinase activity | F |
| ||||||||||||||||||
| GO:0016772 | transferase activity, transferring phosphorus-containing groups | F |
| ||||||||||||||||||
| GO:0017084 | delta1-pyrroline-5-carboxylate synthetase activity | F |
| ||||||||||||||||||
| GO:0006417 | regulation of translation | P |
| ||||||||||||||||||
| GO:0006536 | glutamate metabolic process | P |
| ||||||||||||||||||
| GO:0006561 | proline biosynthetic process | P |
| ||||||||||||||||||
| GO:0006979 | response to oxidative stress | P |
| ||||||||||||||||||
| GO:0008150 | biological_process | P |
| ||||||||||||||||||
| GO:0009269 | response to desiccation | P |
| ||||||||||||||||||
| GO:0009414 | response to water deprivation | P |
| ||||||||||||||||||
| GO:0009651 | response to salt stress | P |
| ||||||||||||||||||
| GO:0009737 | response to abscisic acid stimulus | P |
| ||||||||||||||||||
| GO:0009793 | embryo development ending in seed dormancy | P |
| ||||||||||||||||||
| GO:0018193 | peptidyl-amino acid modification | P |
| ||||||||||||||||||
| GO:0042538 | hyperosmotic salinity response | P |
| ||||||||||||||||||
| GO:0048364 | root development | P |
|
| HomoloGene cluster | Phylogenetic pattern |
| 5683 |  |
| GO ID | GO term | GO aspect | Evidence codes and associations | ||||||||||||||||||
| GO:0005737 | cytoplasm | C |
| ||||||||||||||||||
| GO:0004349 | glutamate 5-kinase activity | F |
| ||||||||||||||||||
| GO:0006417 | regulation of translation | P |
| ||||||||||||||||||
| GO:0006536 | glutamate metabolic process | P |
| ||||||||||||||||||
| GO:0006561 | proline biosynthetic process | P |
| ||||||||||||||||||
| GO:0018193 | peptidyl-amino acid modification | P |
|
| OrthoMCL cluster | Phylogenetic pattern |
| OG1_3324 | 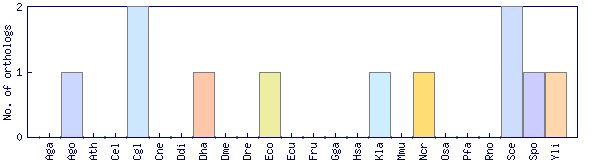 |
| GO ID | GO term | GO aspect | Evidence codes and associations | ||||||||||||||||||
| GO:0005737 | cytoplasm | C |
| ||||||||||||||||||
| GO:0003674 | molecular_function | F |
| ||||||||||||||||||
| GO:0004349 | glutamate 5-kinase activity | F |
| ||||||||||||||||||
| GO:0016301 | kinase activity | F |
| ||||||||||||||||||
| GO:0016772 | transferase activity, transferring phosphorus-containing groups | F |
| ||||||||||||||||||
| GO:0006417 | regulation of translation | P |
| ||||||||||||||||||
| GO:0006536 | glutamate metabolic process | P |
| ||||||||||||||||||
| GO:0006561 | proline biosynthetic process | P |
| ||||||||||||||||||
| GO:0008150 | biological_process | P |
| ||||||||||||||||||
| GO:0018193 | peptidyl-amino acid modification | P |
|
Curated yeast ortholog results:
| S. pombe systematic ID | S. cerevisiae systematic ID |
| SPAC17H9.13c (O13810) | YDR300C (S000002708) |
| GO ID | GO term | GO aspect | Evidence codes and associations | ||||||||||||||||||
| GO:0005737 | cytoplasm | C |
| ||||||||||||||||||
| GO:0004349 | glutamate 5-kinase activity | F |
| ||||||||||||||||||
| GO:0006417 | regulation of translation | P |
| ||||||||||||||||||
| GO:0006536 | glutamate metabolic process | P |
| ||||||||||||||||||
| GO:0006561 | proline biosynthetic process | P |
| ||||||||||||||||||
| GO:0018193 | peptidyl-amino acid modification | P |
|
| YOGY Home | YOGY Help |
| s.khadayate@ucl.ac.uk |