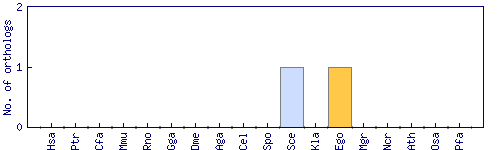
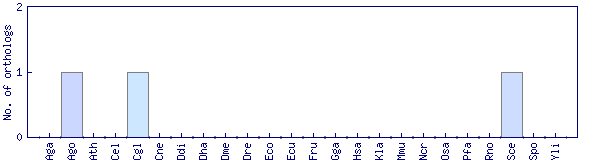
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | RSC3 | ||||||||||
| SGD link | S000002711 | ||||||||||
| Alternative ID | YDR303C | ||||||||||
| Description | Component of the RSC chromatin remodeling complex; essential gene required for maintenance of proper ploidy and regulation of ribosomal protein genes and the cell wall/stress response; highly similar to Rsc30p | ||||||||||
| Synonyms | YDR303C | ||||||||||
| Datasets |
|