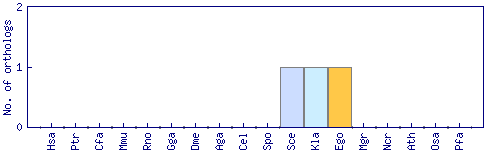
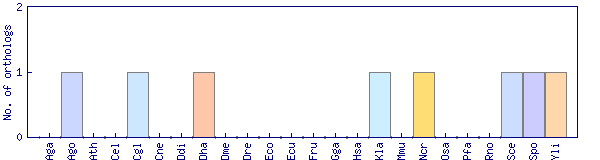
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | IPK1 | ||||||||||
| SGD link | S000002723 | ||||||||||
| Alternative ID | YDR315C | ||||||||||
| Description | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase, nuclear protein required for synthesis of 1,2,3,4,5,6-hexakisphosphate (phytate), which is integral to cell function; has 2 motifs conserved in other fungi; ipk1 gle1 double mutant is inviable | ||||||||||
| Synonyms | YDR315C | ||||||||||
| Datasets |
|