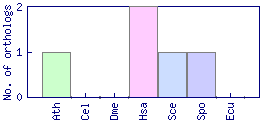
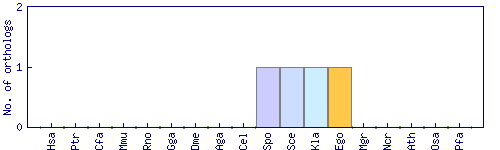
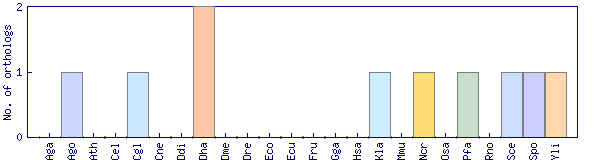
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | OMS1 | ||||||||||
| SGD link | S000002724 | ||||||||||
| Alternative ID | YDR316W | ||||||||||
| Description | Protein integral to the mitochondrial membrane; has a conserved methyltransferase motif; multicopy suppressor of respiratory defects caused by OXA1 mutations | ||||||||||
| Synonyms | YDR316W | ||||||||||
| Datasets |
|