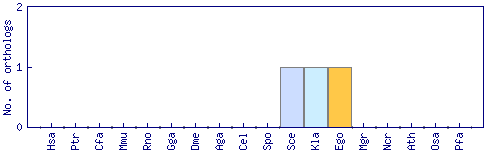
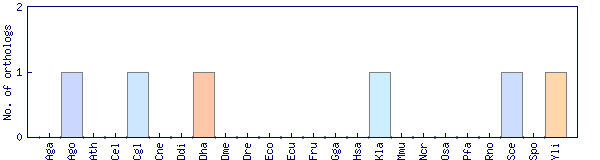
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | ESC2 | ||||||||||
| SGD link | S000002771 | ||||||||||
| Alternative ID | YDR363W | ||||||||||
| Description | Protein involved in silencing; may recruit or stabilize Sir proteins; role in Rad51-dependent homologous recombination repair and intra S-phase DNA damage checkpoint; member of the RENi (Rad60-Esc2-Nip45) family of SUMO-like domain proteins | ||||||||||
| Synonyms | YDR363W | ||||||||||
| Datasets |
|