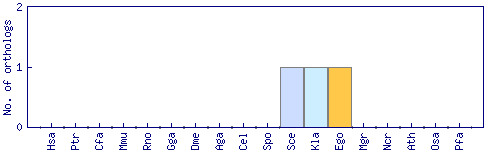
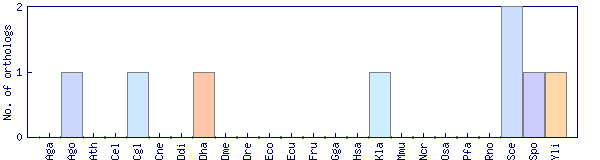
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | PLM2 | ||||||||||
| SGD link | S000002909 | ||||||||||
| Alternative ID | YDR501W | ||||||||||
| Description | Forkhead Associated domain containing protein and putative transcription factor found associated with chromatin; target of SBF transcription factor; induced in response to DNA damaging agents and deletion of telomerase; similar to TOS4 | ||||||||||
| Synonyms | YDR501W | ||||||||||
| Datasets |
|