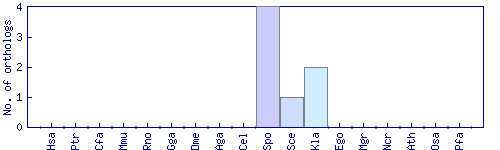
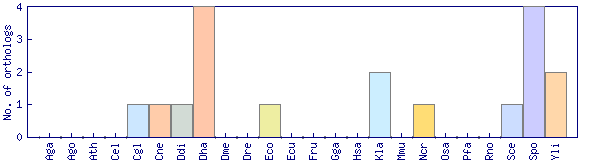
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | HSP31 | ||||||||||
| SGD link | S000002941 | ||||||||||
| Alternative ID | YDR533C | ||||||||||
| Description | Possible chaperone and cysteine protease with similarity to E. coli Hsp31; member of the DJ-1/ThiJ/PfpI superfamily, which includes human DJ-1 involved in Parkinson's disease; exists as a dimer and contains a putative metal-binding site | ||||||||||
| Synonyms | YDR533C | ||||||||||
| Datasets |
|