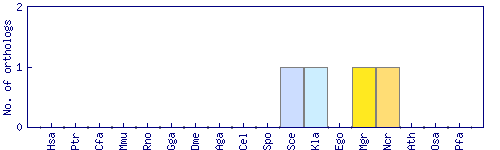
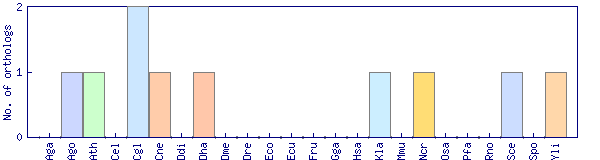
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | UTR2 | ||||||||||
| SGD link | S000000766 | ||||||||||
| Alternative ID | YEL040W | ||||||||||
| Description | Chitin transglycosylase that functions in the transfer of chitin to beta(1-6) and beta(1-3) glucans in the cell wall; similar to and functionally redundant with Crh1; glycosylphosphatidylinositol (GPI)-anchored protein localized to bud neck | ||||||||||
| Synonyms | YEL040W | ||||||||||
| Datasets |
|