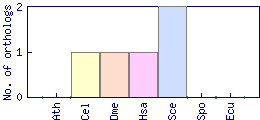
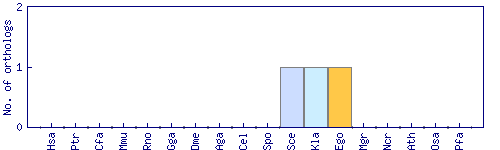
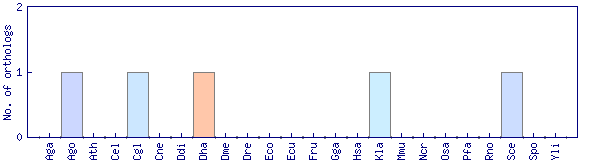
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | YAT2 | ||||||||||
| SGD link | S000000826 | ||||||||||
| Alternative ID | YER024W | ||||||||||
| Description | Carnitine acetyltransferase; has similarity to Yat1p, which is a carnitine acetyltransferase associated with the mitochondrial outer membrane | ||||||||||
| Synonyms | YER024W | ||||||||||
| Datasets |
|