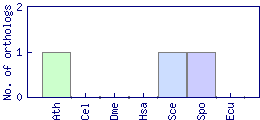
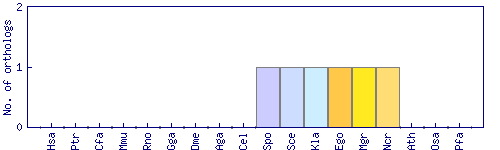
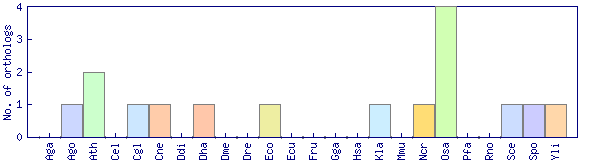
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | HIS1 | ||||||||||
| SGD link | S000000857 | ||||||||||
| Alternative ID | YER055C | ||||||||||
| Description | ATP phosphoribosyltransferase, a hexameric enzyme, catalyzes the first step in histidine biosynthesis; mutations cause histidine auxotrophy and sensitivity to Cu, Co, and Ni salts; transcription is regulated by general amino acid control | ||||||||||
| Synonyms | YER055C | ||||||||||
| Datasets |
|