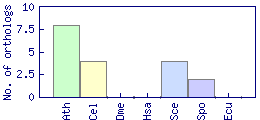
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | ICL1 | ||||||||||
| SGD link | S000000867 | ||||||||||
| Alternative ID | YER065C | ||||||||||
| Description | Isocitrate lyase, catalyzes the formation of succinate and glyoxylate from isocitrate, a key reaction of the glyoxylate cycle; expression of ICL1 is induced by growth on ethanol and repressed by growth on glucose | ||||||||||
| Synonyms | YER065C | ||||||||||
| Datasets |
|