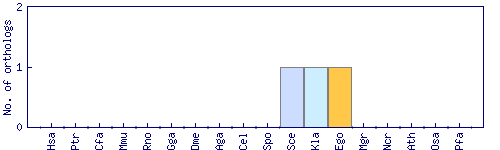
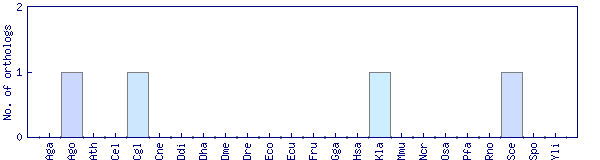
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | SCC4 | ||||||||||
| SGD link | S000000949 | ||||||||||
| Alternative ID | YER147C | ||||||||||
| Description | Subunit of cohesin loading factor (Scc2p-Scc4p), a complex required for the loading of cohesin complexes onto chromosomes; involved in establishing sister chromatid cohesion during double-strand break repair via phosphorylated histone H2AX | ||||||||||
| Synonyms | YER147C | ||||||||||
| Datasets |
|