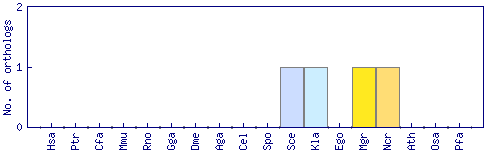
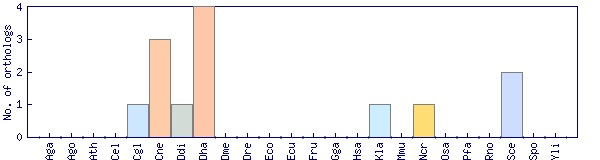
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | PUG1 | ||||||||||
| SGD link | S000000987 | ||||||||||
| Alternative ID | YER185W | ||||||||||
| Description | Plasma membrane protein with roles in the uptake of protoprophyrin IX and the efflux of heme; expression is induced under both low-heme and low-oxygen conditions; member of the fungal lipid-translocating exporter (LTE) family of proteins | ||||||||||
| Synonyms | YER185W | ||||||||||
| Datasets |
|