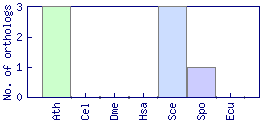
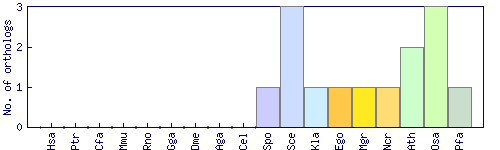
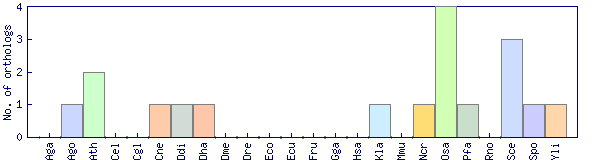
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | SNZ3 | ||||||||||
| SGD link | S000001835 | ||||||||||
| Alternative ID | YFL059W | ||||||||||
| Description | Member of a stationary phase-induced gene family; transcription of SNZ2 is induced prior to diauxic shift, and also in the absence of thiamin in a Thi2p-dependent manner; forms a coregulated gene pair with SNO3 | ||||||||||
| Synonyms | YFL059W | ||||||||||
| Datasets |
|