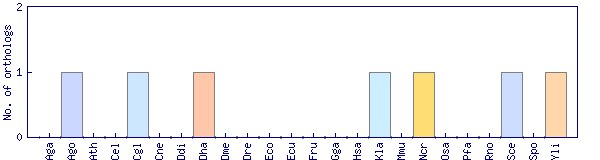
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | PHO4 | ||||||||||
| SGD link | S000001930 | ||||||||||
| Alternative ID | YFR034C | ||||||||||
| Description | Basic helix-loop-helix (bHLH) transcription factor of the myc-family; binds cooperatively with Pho2p to the PHO5 promoter; function is regulated by phosphorylation at multiple sites and by phosphate availability | ||||||||||
| Synonyms | YFR034C | ||||||||||
| Datasets |
|