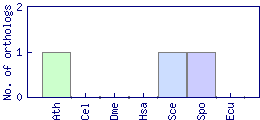
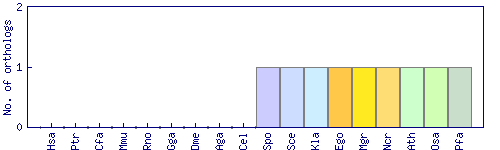
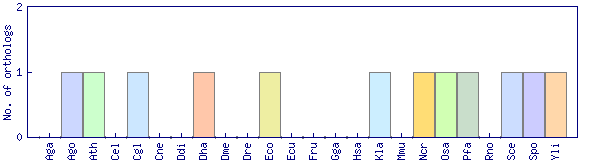
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | ARO2 | ||||||||||
| SGD link | S000003116 | ||||||||||
| Alternative ID | YGL148W | ||||||||||
| Description | Bifunctional chorismate synthase and flavin reductase, catalyzes the conversion of 5-enolpyruvylshikimate 3-phosphate (EPSP) to form chorismate, which is a precursor to aromatic amino acids | ||||||||||
| Synonyms | YGL148W | ||||||||||
| Datasets |
|