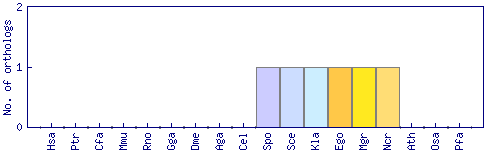
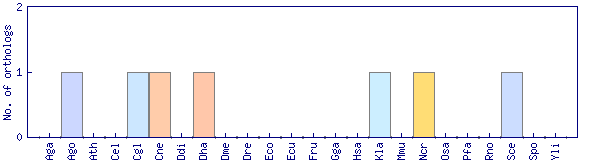
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | GTS1 | ||||||||||
| SGD link | S000003149 | ||||||||||
| Alternative ID | YGL181W | ||||||||||
| Description | Arf3p GTPase Activating Protein (GAP) that localizes to endocytic patches; gts1 mutations affect budding, cell size, heat tolerance, sporulation, life span, ultradian rhythms; localizes to nucleus and induces flocculation when overexpressed | ||||||||||
| Synonyms | YGL181W | ||||||||||
| Datasets |
|