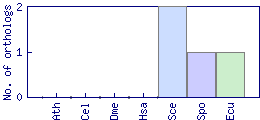
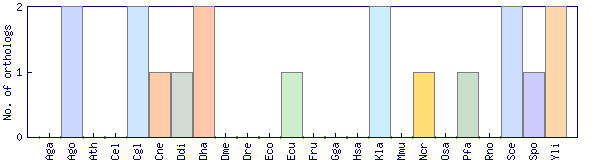
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | BRR6 | ||||||||||
| SGD link | S000003216 | ||||||||||
| Alternative ID | YGL247W | ||||||||||
| Description | Essential nuclear envelope integral membrane protein required for nuclear envelope morphology, nuclear pore complex localization, nuclear export; exhibits synthetic lethal genetic interactions with genes involved in lipid metabolism | ||||||||||
| Synonyms | YGL247W | ||||||||||
| Datasets |
|