| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | SKN1 | ||||||||||
| SGD link | S000003375 | ||||||||||
| Alternative ID | YGR143W | ||||||||||
| Description | Protein involved in sphingolipid biosynthesis; type II membrane protein with similarity to Kre6p | ||||||||||
| Synonyms | YGR143W | ||||||||||
| Datasets |
|
| YOGY Home | YOGY Help |
|
| HomoloGene cluster | Phylogenetic pattern |
| 40554 | 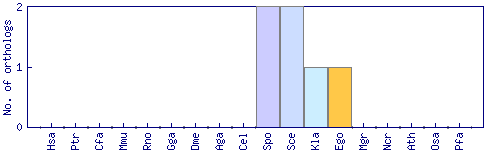 |
| GO ID | GO term | GO aspect | Evidence codes and associations | ||||||||||
| GO:0000137 | Golgi cis cisterna | C |
| ||||||||||
| GO:0005783 | endoplasmic reticulum | C |
| ||||||||||
| GO:0016021 | integral to membrane | C |
| ||||||||||
| GO:0030176 | integral to endoplasmic reticulum membrane | C |
| ||||||||||
| GO:0015926 | glucosidase activity | F |
| ||||||||||
| GO:0006078 | 1,6-beta-glucan biosynthetic process | P |
| ||||||||||
| GO:0007047 | cellular cell wall organization | P |
| ||||||||||
| GO:0030148 | sphingolipid biosynthetic process | P |
| ||||||||||
| GO:0031505 | fungal-type cell wall organization | P |
|
| OrthoMCL cluster | Phylogenetic pattern |
| OG1_4321 | 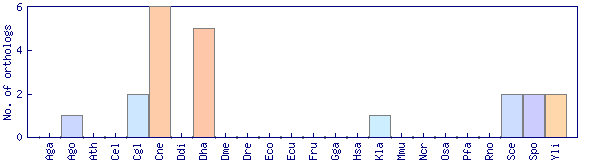 |
| GO ID | GO term | GO aspect | Evidence codes and associations | ||||||||||
| GO:0000137 | Golgi cis cisterna | C |
| ||||||||||
| GO:0005783 | endoplasmic reticulum | C |
| ||||||||||
| GO:0016021 | integral to membrane | C |
| ||||||||||
| GO:0030176 | integral to endoplasmic reticulum membrane | C |
| ||||||||||
| GO:0015926 | glucosidase activity | F |
| ||||||||||
| GO:0006078 | 1,6-beta-glucan biosynthetic process | P |
| ||||||||||
| GO:0007047 | cellular cell wall organization | P |
| ||||||||||
| GO:0030148 | sphingolipid biosynthetic process | P |
| ||||||||||
| GO:0031505 | fungal-type cell wall organization | P |
|
Curated yeast ortholog results:
| S. pombe systematic ID | S. cerevisiae systematic ID |
| SPAC23H3.11c (O13941) | YGR143W (S000003375) |
| SPAC17G6.11c (O13789) | YGR143W (S000003375) |
| GO ID | GO term | GO aspect | Evidence codes and associations | ||||||||||
| GO:0005783 | endoplasmic reticulum | C |
| ||||||||||
| GO:0016021 | integral to membrane | C |
| ||||||||||
| GO:0015926 | glucosidase activity | F |
| ||||||||||
| GO:0006078 | 1,6-beta-glucan biosynthetic process | P |
| ||||||||||
| GO:0007047 | cellular cell wall organization | P |
| ||||||||||
| GO:0030148 | sphingolipid biosynthetic process | P |
| ||||||||||
| GO:0031505 | fungal-type cell wall organization | P |
|
| YOGY Home | YOGY Help |
| s.khadayate@ucl.ac.uk |