| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | BGL2 | ||||||||||
| SGD link | S000003514 | ||||||||||
| Alternative ID | YGR282C | ||||||||||
| Description | Endo-beta-1,3-glucanase, major protein of the cell wall, involved in cell wall maintenance | ||||||||||
| Synonyms | YGR282C | ||||||||||
| Datasets |
|
| YOGY Home | YOGY Help |
|
| HomoloGene cluster | Phylogenetic pattern |
| 39184 | 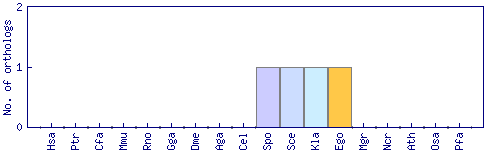 |
| GO ID | GO term | GO aspect | Evidence codes and associations | ||||||||
| GO:0009277 | fungal-type cell wall | C |
| ||||||||
| GO:0004338 | glucan 1,3-beta-glucosidase activity | F |
| ||||||||
| GO:0042124 | 1,3-beta-glucanosyltransferase activity | F |
| ||||||||
| GO:0042973 | glucan endo-1,3-beta-D-glucosidase activity | F |
| ||||||||
| GO:0006076 | 1,3-beta-glucan catabolic process | P |
| ||||||||
| GO:0006414 | translational elongation | P |
| ||||||||
| GO:0007047 | cellular cell wall organization | P |
| ||||||||
| GO:0031505 | fungal-type cell wall organization | P |
| ||||||||
| GO:0044262 | cellular carbohydrate metabolic process | P |
|
| OrthoMCL cluster | Phylogenetic pattern |
| OG1_11603 | 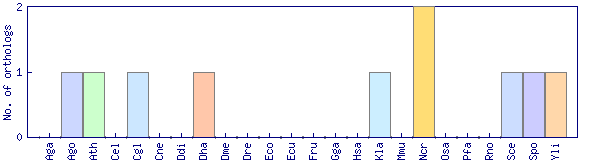 |
| Species | OrthoMCL sequence | Model organism protein page | UniProt link |
| Ashbya gossypii (Ago) | ago3969 | AGL343C | |
| Arabidopsis thaliana (Ath) | ath17624 | At3g61810.1 | |
| Candida glabrata (Cgl) | cgl1635 | CAGL0G00220g | |
| Debaryomyces hansenii (Dha) | dha6591 | DEHA0G19943g | |
| Kluyveromyces lactis (Kla) | kla4217 | KLLA0F03036g | |
| Neurospora crassa (Ncr) | ncr6200 | NCU06381.2 | |
| Neurospora crassa (Ncr) | ncr8942 | NCU09175.2 | |
| Saccharomyces cerevisiae (Sce) | sce2666 | YGR282C (S000003514) | |
| Schizosaccharomyces pombe (Spo) | spo2775 | SPAC26H5.08c | O13990 |
| Yarrowia lipolytica (Yli) | yli893 | YALI0B03564g |
| GO ID | GO term | GO aspect | Evidence codes and associations | ||||||||
| GO:0009277 | fungal-type cell wall | C |
| ||||||||
| GO:0004338 | glucan 1,3-beta-glucosidase activity | F |
| ||||||||
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds | F |
| ||||||||
| GO:0042124 | 1,3-beta-glucanosyltransferase activity | F |
| ||||||||
| GO:0042973 | glucan endo-1,3-beta-D-glucosidase activity | F |
| ||||||||
| GO:0006076 | 1,3-beta-glucan catabolic process | P |
| ||||||||
| GO:0006414 | translational elongation | P |
| ||||||||
| GO:0007047 | cellular cell wall organization | P |
| ||||||||
| GO:0031505 | fungal-type cell wall organization | P |
| ||||||||
| GO:0044262 | cellular carbohydrate metabolic process | P |
|
Curated yeast ortholog results:
| S. pombe systematic ID | S. cerevisiae systematic ID |
| SPAC26H5.08c (O13990) | YGR282C (S000003514) |
| GO ID | GO term | GO aspect | Evidence codes and associations | ||||||||
| GO:0009277 | fungal-type cell wall | C |
| ||||||||
| GO:0004338 | glucan 1,3-beta-glucosidase activity | F |
| ||||||||
| GO:0042124 | 1,3-beta-glucanosyltransferase activity | F |
| ||||||||
| GO:0042973 | glucan endo-1,3-beta-D-glucosidase activity | F |
| ||||||||
| GO:0006076 | 1,3-beta-glucan catabolic process | P |
| ||||||||
| GO:0006414 | translational elongation | P |
| ||||||||
| GO:0007047 | cellular cell wall organization | P |
| ||||||||
| GO:0031505 | fungal-type cell wall organization | P |
| ||||||||
| GO:0044262 | cellular carbohydrate metabolic process | P |
|
| YOGY Home | YOGY Help |
| s.khadayate@ucl.ac.uk |