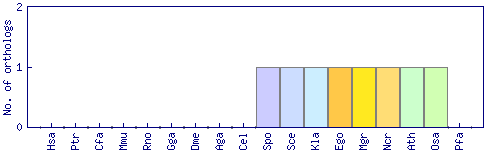
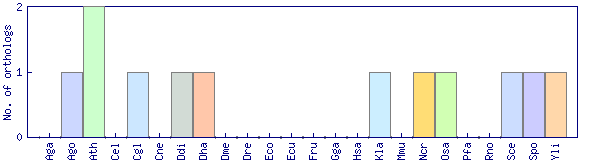
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | MSC7 | ||||||||||
| SGD link | S000001081 | ||||||||||
| Alternative ID | YHR039C | ||||||||||
| Description | Protein of unknown function, green fluorescent protein (GFP)-fusion protein localizes to the endoplasmic reticulum; msc7 mutants are defective in directing meiotic recombination events to homologous chromatids | ||||||||||
| Synonyms | YHR039C | ||||||||||
| Datasets |
|