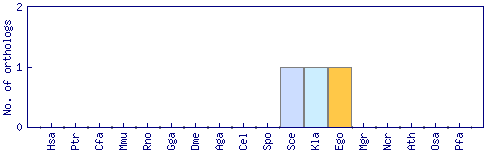
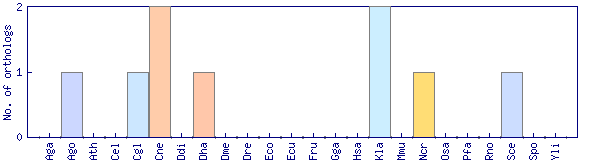
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | STB5 | ||||||||||
| SGD link | S000001221 | ||||||||||
| Alternative ID | YHR178W | ||||||||||
| Description | Transcription factor, involved in regulating multidrug resistance and oxidative stress response; forms a heterodimer with Pdr1p; contains a Zn(II)2Cys6 zinc finger domain that interacts with a pleiotropic drug resistance element in vitro | ||||||||||
| Synonyms | YHR178W | ||||||||||
| Datasets |
|