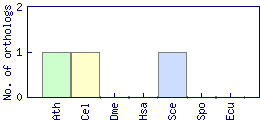
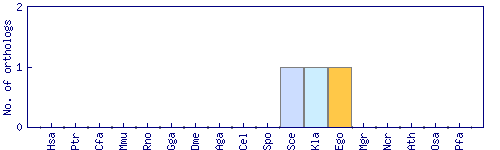
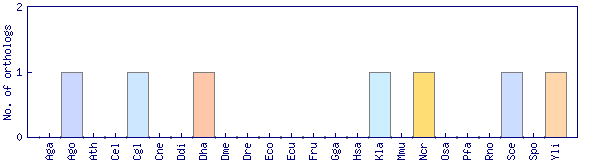
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | CTF8 | ||||||||||
| SGD link | S000001234 | ||||||||||
| Alternative ID | YHR191C | ||||||||||
| Description | Subunit of a complex with Ctf18p that shares some subunits with Replication Factor C and is required for sister chromatid cohesion | ||||||||||
| Synonyms | YHR191C | ||||||||||
| Datasets |
|