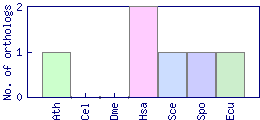
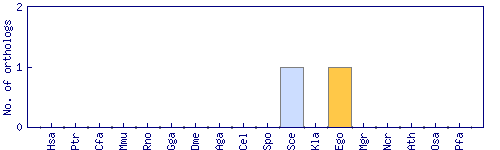
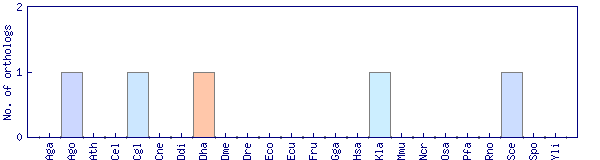
| Species | Saccharomyces cerevisiae (Sce) | ||||||||||
| Primary name | VID28 | ||||||||||
| SGD link | S000001279 | ||||||||||
| Alternative ID | YIL017C | ||||||||||
| Description | Protein involved in proteasome-dependent catabolite degradation of fructose-1,6-bisphosphatase (FBPase); localized to the nucleus and the cytoplasm | ||||||||||
| Synonyms | YIL017C | ||||||||||
| Datasets |
|